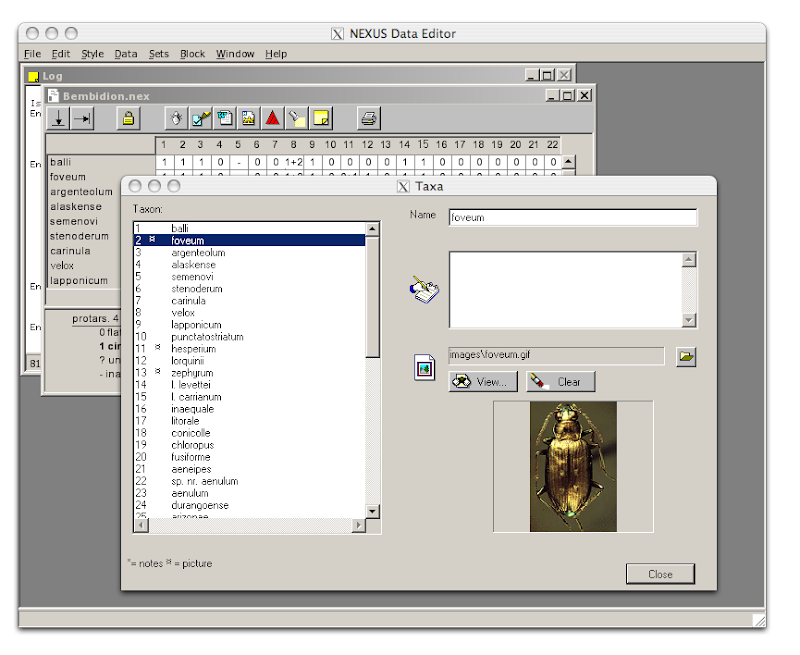
One of the potentially powerful features of TreeBASE II is availability of a RDF version of a study. This means that, in principle, one could take the RDF for a TreeBASE study, combine it with RDF from other sources, and generate a richer view of a particular study. For example, if a TreeBASE study has a DOI, then we could link it to bibliographic details for the study, and through them to other information, such as GenBank sequences, specimens, etc. (see my
little linked data browser for an example of some of this linking). If we added a phylogeny viewer, then we'd have a great tool for browsing the basic components of a phylogenetic study.
Unfortunately, we're not there yet. I've been trying to make sense of TreeBASE II RDF, and frankly, it's a mess. Here are some of the problems:
TreeBASE URIs aren't linked data compliantThe canonical URI for a study (e.g.,
http://purl.org/phylo/treebase/phylows/study/TB2:S10423) doesn't conform to the linked data approach. In fact, the URI crashes the
linked data validator, so I tried another test.
curl --include
--header "Accept: application/rdf+xml"
http://purl.org/phylo/treebase/phylows/study/TB2:S10423
To be a valid linked data resource this request should return a 303 HTTP status code. Instead we get a 302 and some HTML. Linked data clients won't be able to extract information from this URI.
SKOS matchingThere are some odd things going on in the RDF. It contains statements of the form:
<rdf:Description rdf:ID="otu1789319">
<skos:closeMatch rdf:resource="http://purl.uniprot.org/taxonomy/76066.rdf">
</rdf:Description>
(I've tidied this up a little from the original, rather verbose RDF). This asserts that the TreeBASE OTU
otu1789319 corresponds to the NCBI taxon with the taxonomy id
76066 (represented by the Uniprot URI). Except, it doesn't really. As far as I understand it, SKOS is about matching concepts, not documents. The URI
http://purl.uniprot.org/taxonomy/76066.rdf is a document URI (specifically, a RDF document), the URI
http://purl.uniprot.org/taxonomy/76066 is the taxon. The match should really be to http://purl.uniprot.org/taxonomy/76066. Then I've come across statements that match TreeBASE OTUs to
http://purl.uniprot.org/taxonomy/0.rdf. This URI doesn't exist (we get a 404). This seems an odd way to say that we don't have a match -- if we don't have a match, don't include it in the RDF.
Local URIs for trees don't workThe RDF is full of local URIs such as
http://purl.org/phylo/treebase/phylows/#tree1790755, which don't resolve. In fact they generate a rather spectacular Tomcat exception. I don't understand why we need local URIs.
Everything in TreeBASE should have a global URI. Then we can avoid unnecessary statements such as:
http://purl.org/phylo/treebase/phylows/#tree1790755 owl:sameAs http://purl.org/phylo/treebase/phylows/tree/TB2:Tr7899
which links a local resource to a global one
http://purl.org/phylo/treebase/phylows/tree/TB2:Tr7899. Incidentally, this URI doesn't resolve, despite claims that
this bug has been fixed.
No links between tree and studyBut the show stopper for me is that there is
no link between a study and a tree! There is no triple in the RDF specifying any relationship between these two entities. To me this is just about the most important thing I need. I want to be able to query TreeBASE RDF using a study identifier (either from TreeBASE itself, or from an external identifier such as a DOI or a PubMed number). As it stands the TreeBASE II RDF is almost useless. I can't get it via a linked data client, it's full of URIs that don't resolve, and it lacks key triples that would glue things together.
RDF != XMLI can't help thinking that the RDF output hasn't been designed with end use in mind. I know from my own experience that it's not until you try to do something with the RDF that you realise how poor some design decisions may have been.
It's not enough to pump out RDF and hope for the best. RDF is not XML, which is just a verbose format for moving data around. RDF brings with it all sorts of expectations about how clients will resolve it, how they will interpret URIs, and the kinds of queries that will be performed. We are achingly close to being able to tie everything together, but not with RDF TreeBASE II is currently making available.


 In an
In an