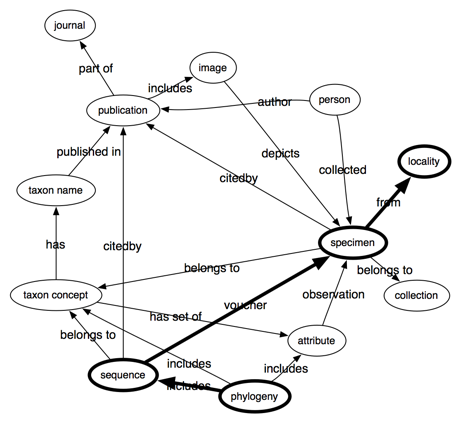
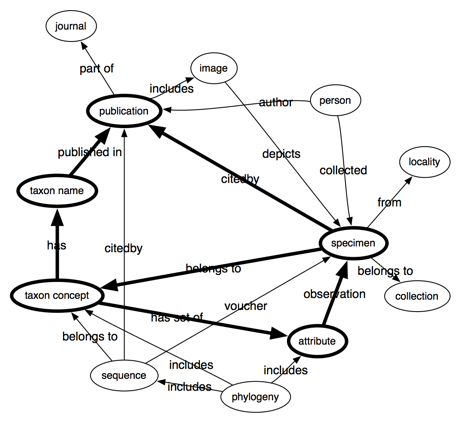
Following on from my
previous post bemoaning the lack of links between biodiversity data sets, it's worth looking at different ways we can build these links. Specifically, data can be tightly or loosely coupled.
Tight coupling
Tight coupling uses identifiers. A good example is bibliographic citation, where we state that one reference cites another by linking DOIs. This makes it easy to store these links in a database, such as the
Open Citations project which is exploring citation networks base don data from PubMed Cenral. Tight coupling also makes it easy to aggregate information from multiple sources. For example, one database may record citations of a paper, another may record citations of GenBank sequences, a third may record publication of taxonomic names. If all three databases use the same identifiers for the same publications (e.g., DOIs) we can combine them and potentially discover new things (for example, we could answer the question "how many descriptions of new species include sequence data?").
Loose coupling
In part this post has been prompted by a
discussion I've been having with Paul Murray (
@PaulMurrayCbr on his blog. Paul has added
COinS to pages in the
Australian Faunal Directory (AFD). These are snippets of HTML that encode a bibliographic reference as an OpenURL, and which browser extensions such as
OpenURL Referrer for Firefox and
COinS 2 OpenURL for Chrome can convert into links.
I've mapped many of the references in AFD to standard identifiers such as DOIs, or to digital libraries such as
BioStor, and this tightly-coupled mapping is available in
AFD on CouchDB. To date these mappings haven't been imported into AFD itself, which means that users of the original site don't have easy access to the literature that appears on that site (basically they'll have to Google each reference). However, if they have a browser extension (or the Javascript bookmarklet available from
http://iphylo.org/~rpage/afd/openurl) that supports COinS, they will now see a clickable link that, in many cases, will take them to the online version of the corresponding reference.
This is an example of loose linking. The AFD site provides OpenURL links which can be resolved
"just in time". Users of the AFD site can get some of the benefits of the tight linking stored in my CouchDB version of AFD, but the maintainers of AFD itself don't need to add code to handle these identifiers.
A lot of linking of biodiversity data shares this pattern. Instead of linking identifiers, one site links to another through a query. For example, NCBI taxonomy links to GBIF using URLs of the form "http://data.gbif.org/search/<taxon name>". Linking by query is potentially more robust than simply linking by URLs, especially if the target of the link doesn't ensure its identifiers are stable (GBIF, I'm looking at you). But there may be multiple ways to construct the same search query, which makes them poor candidates for use as identifiers. COinS are perhaps an extreme example, where there are at least two versions of the OpenURL standard in the wild, and the key-value pairs that make up the query can be in any order.
If the goal is to integrate data then having the same identifiers for the same thing make life a lot simpler, and means that we can switch from endless data cleaning and matching ("is this citation the same as that one?") to building systems that can tackle some of the scientific questions we are interested in. But in their absence we are left a kind of
defensive programming where we expect the links to fail. Loose linking creates "soft links" that may work for humans (we get to click on a link and, with luck, see a web page) but they are less useful for mechanised tools trying to aggregate data.
When tight=loose
Although I've distinguished between tight and loose coupling, the distinction is not absolute. Indeed, one could argue that the best "tight" coupling is a form of "loose" coupling. For example, the most obvious form of tight linking is to use URLs for the things of interest. This is simple and direct, but has draw backs for both publisher and consumer. For the consumer, we are now at the mercy of the publisher's ability to keep the URLs stable. If they change (for example, publishing firm is bought by another firm, or adopts new publishing platform which generates different URLs) then the links break (not to mention that URLs for some resources, such as articles, are often conditional on how you are accessing the article, and may contain extraneous cruff such as session ids, etc.).
Likewise, the publisher is now constrained by a decision it made at the time of publication. If it decides to adopt better technology, or if circumstances otherwise change, it may find itself having to break existing identifiers. Some of this can be avoided if we designed clean URLs, such as this example
http://data.rbge.org.uk/herb/E00001195 given by
Roger Hyam. However, I wonder how persistent the ".uk" part of this URL will be if the Royal Botanic Garden Edinburgh finds itself in a Scotland that
is no longer part of the United Kingdom.
One solution is our old friend
indirection, where we put an identifier in between the consumer and the actual URL of the resource, and the consumer uses that identifier. This is the rationale for DOIs. The user gets an identifier that is unlikely to change, and hence can build systems upon that identifier. The publisher knows that they can change how they serve the corresponding data without disrupting their users, so long as they update the URL that the DOI points to. Indirection gives users the appearance of tight coupling without imposing the constraints of tight coupling on publishers.
 The
The 
